New Feature: Annotate the HLA-binding preference of your TCRs with ImmuneWatch DETECT
We have just released a new version of ImmuneWatch DETECT with two major updates:
- New feature: HLA annotations of your TCRs
- Quarterly database release: improved epitope coverage.

New feature: HLA annotations of your TCRs
Several key publications have shown that it is possible to infer HLA alleles directly from a TCR repertoire, indicating that some information about the HLA-binding preference is stored in the T-cell receptor sequence.
Leveraging our unique database of TCR-epitope pairs, we developed an algorithm to see if a query TCR matches a set of known HLA-specific TCRs in the database. The database matching outputs a binding score ranging from 0 (no binding) to 1 (strongest binding) that indicates the likelihood of the TCR interacting with the predicted MHC allele.
We have evaluated the performance by performing HLA-typing on a set of TCR repertoires with known HLA alleles. In this walkthrough, we demonstrated that DETECT can reliably distinguish between HLA-positive and HLA-negative individuals, achieving an AUC of 0.93 at the optimal score threshold of 0.3
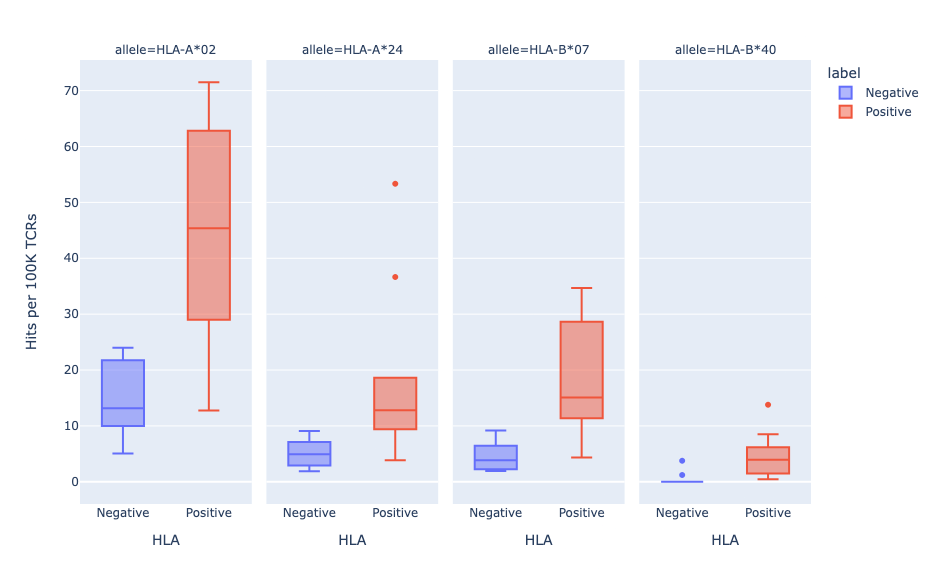
In our latest version (1.4.0) HLA annotations are added to ImmuneWatch DETECT output by default.
Quarterly database release
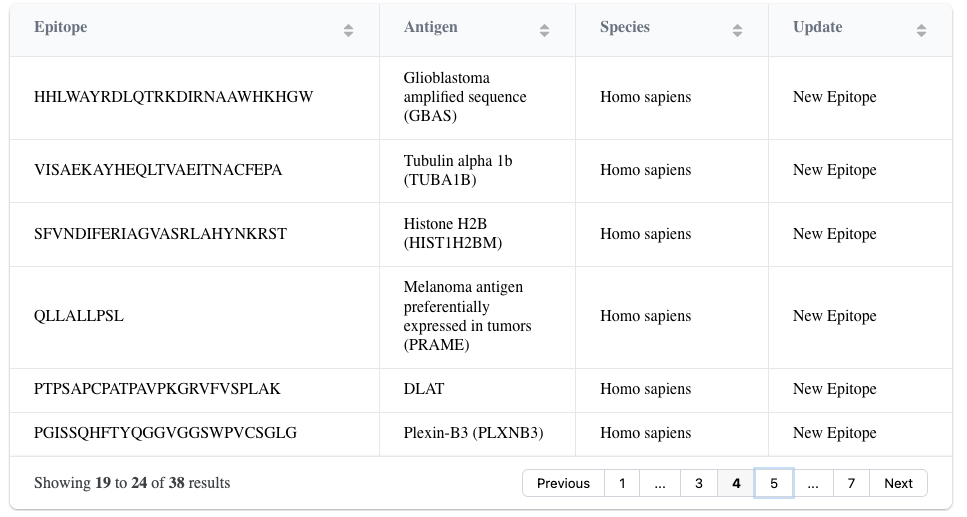
In our database update on July 25, 2025, with ImmuneWatch DETECT Version 1.4.0, we added 1522 new epitope-specific TCRs for 39 different epitopes to our database. Most of the data comes from cancer-specific epitopes.
Get access to both updates by downloading the latest version
Follow the installation instructions described in our documentation pages to get access. It’s as easy as running a one-line-command.
Stay up to date
Subscribe to our newsletter to receive useful tips and information.